Python code for abnormal detection or fault detection using Support Vector Data Description (SVDD)
Python code for abnormal detection or fault detection using Support Vector Data Description (SVDD)
Version 1.1, 11-NOV-2021
Email: iqiukp@outlook.com






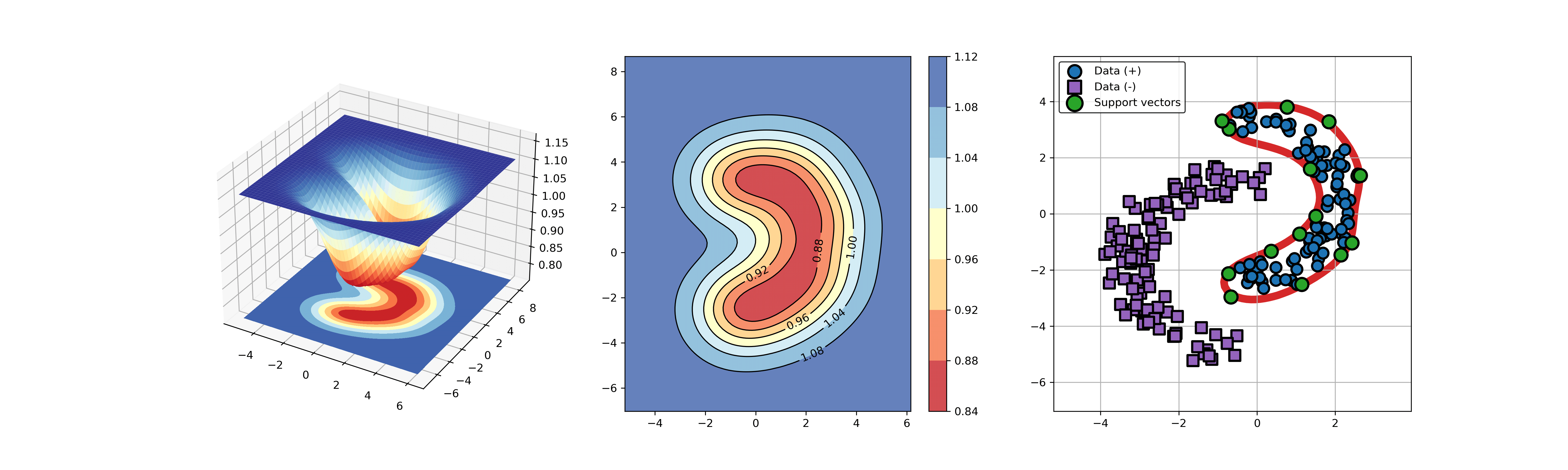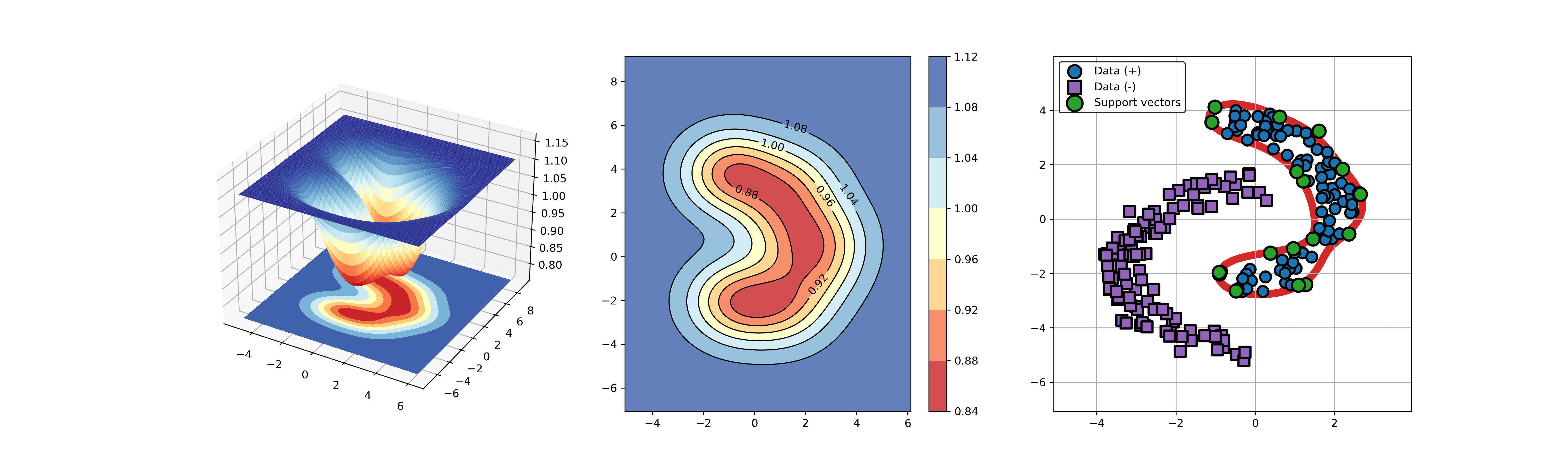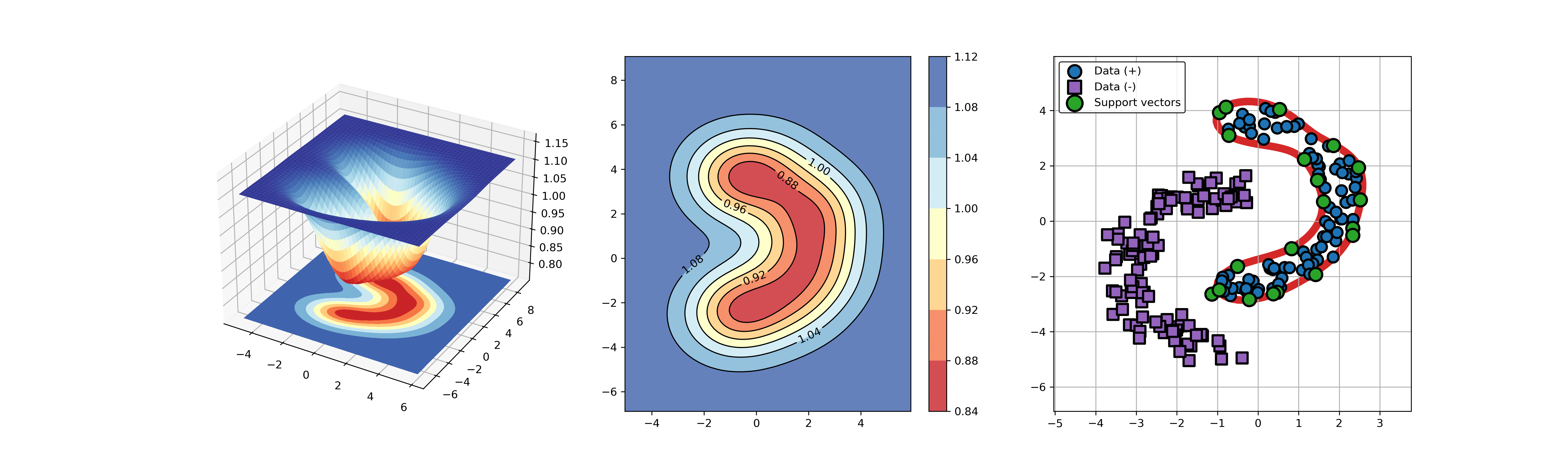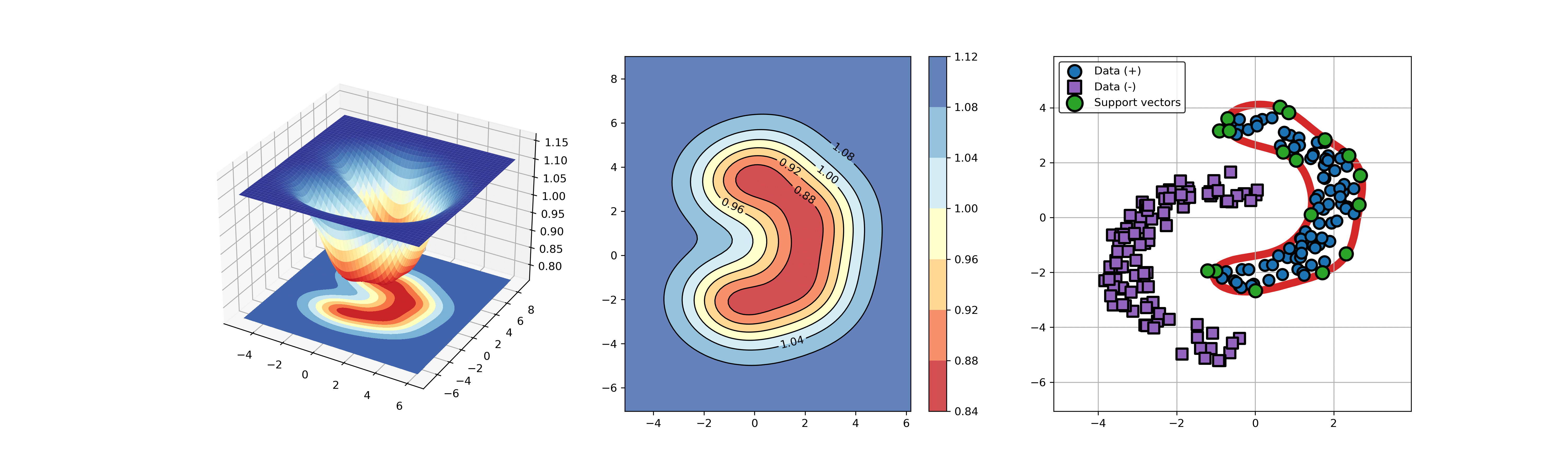
An example for SVDD model fitting using unlabeled data.
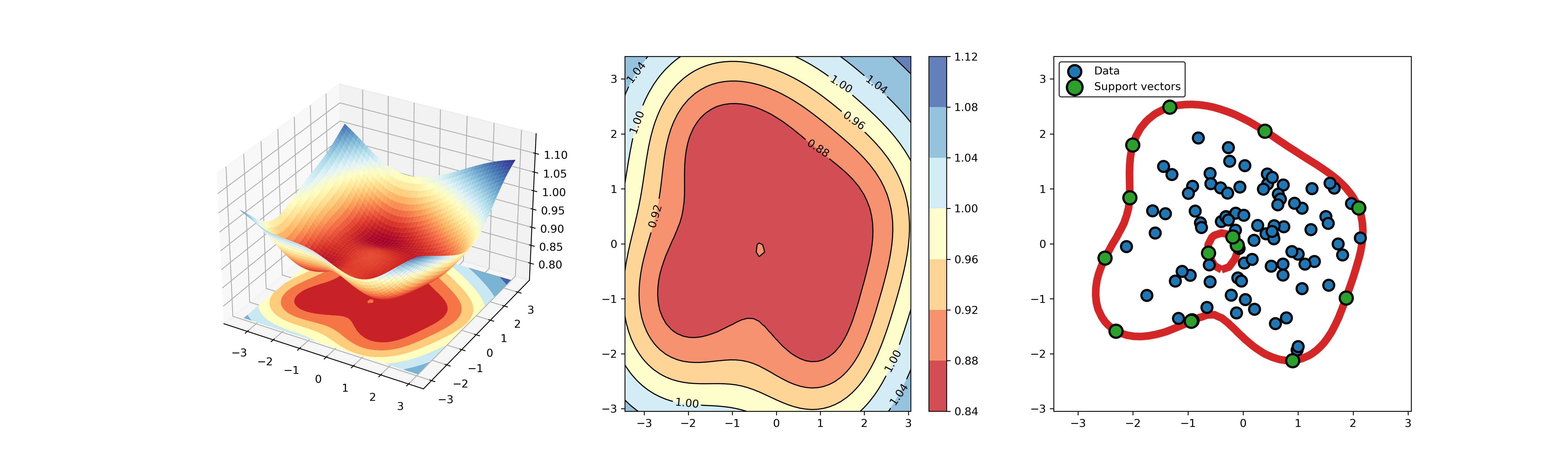
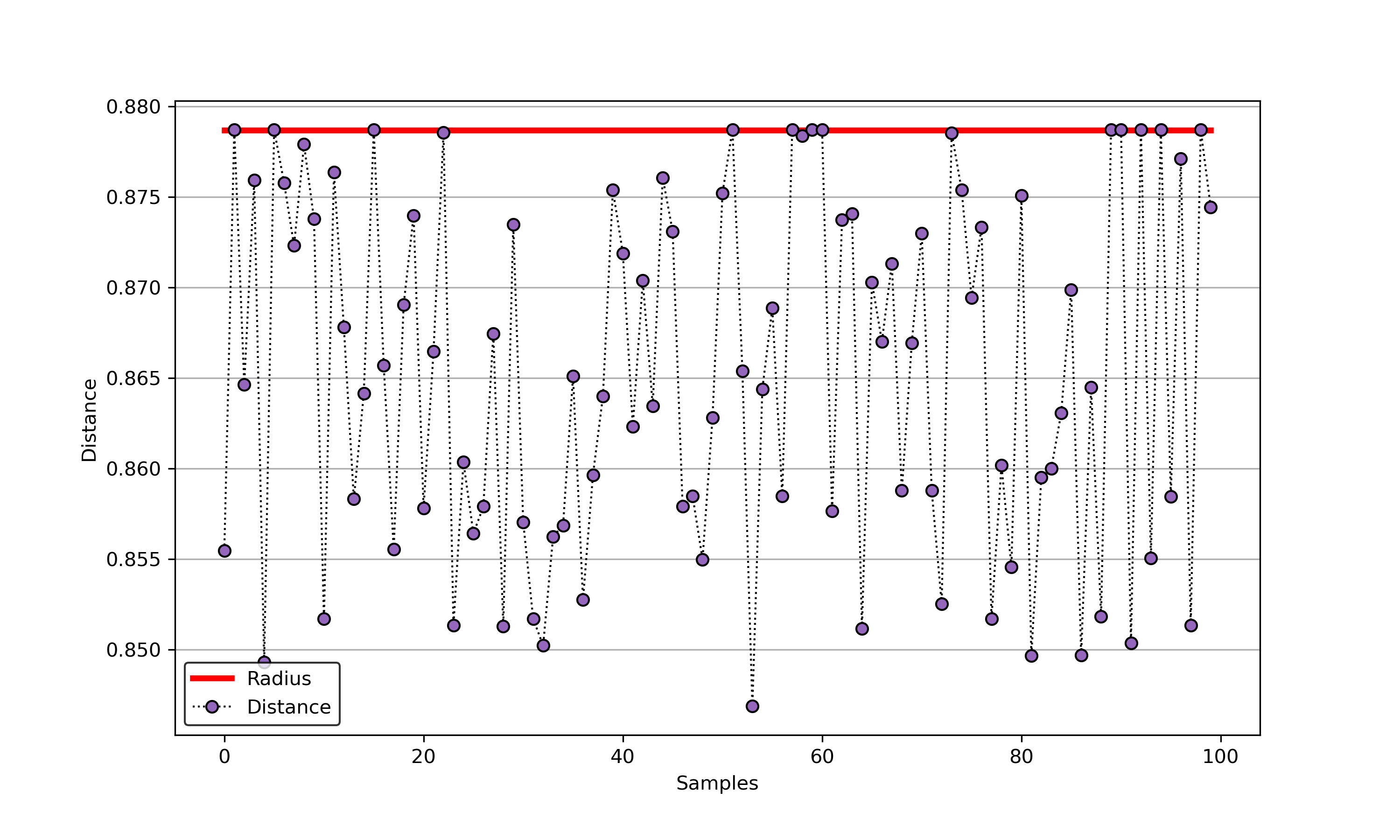
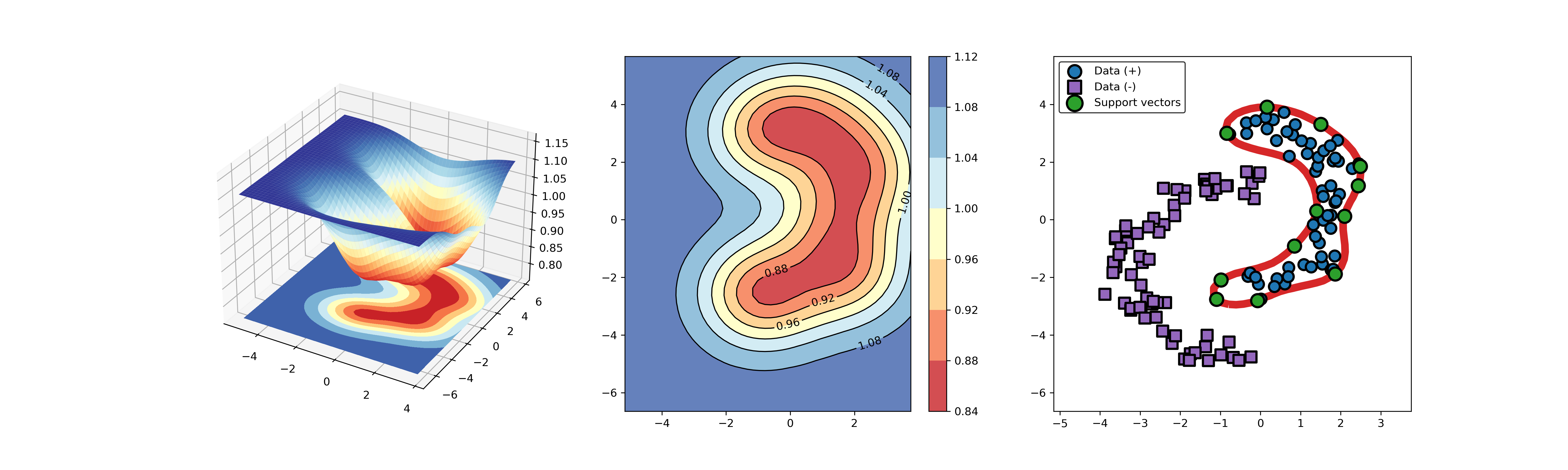
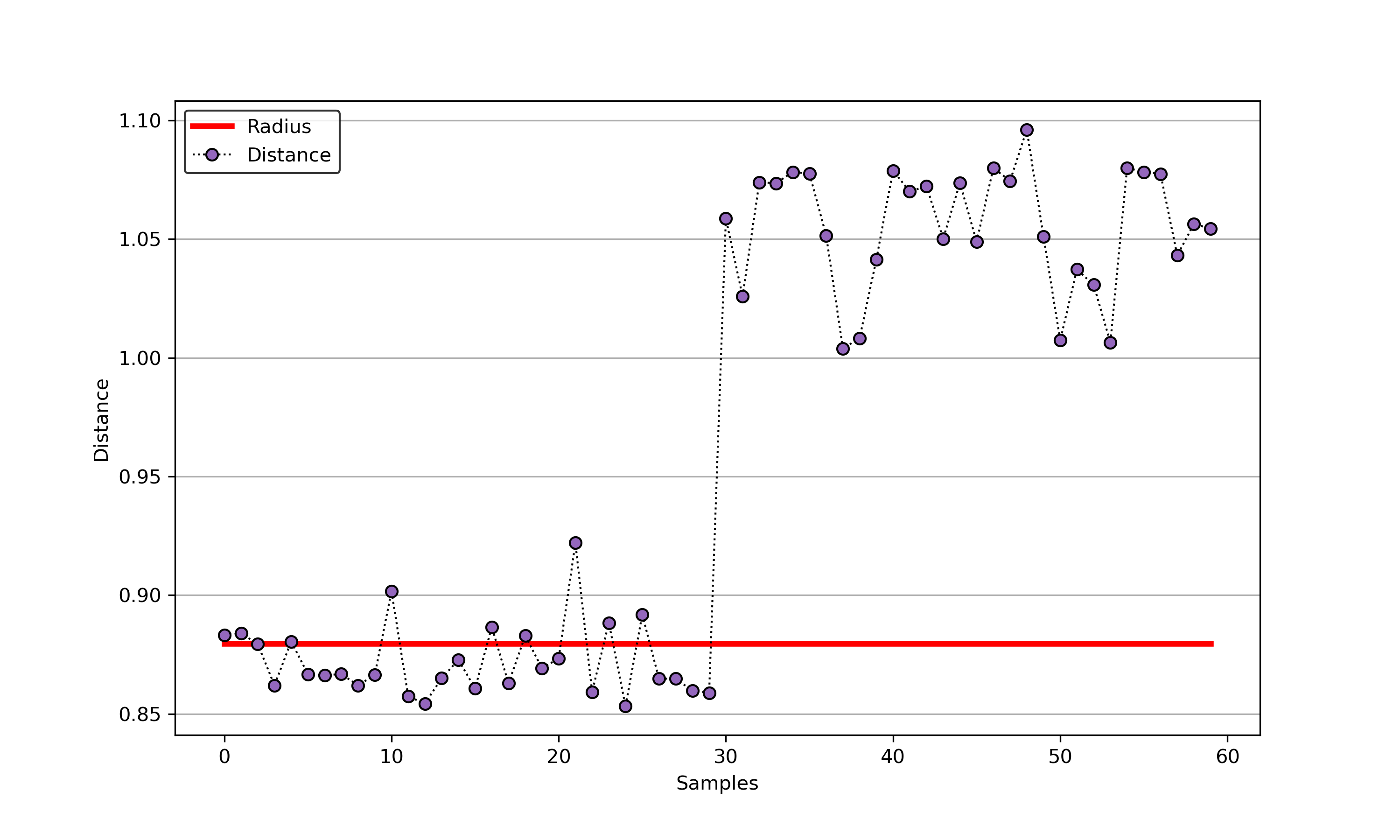
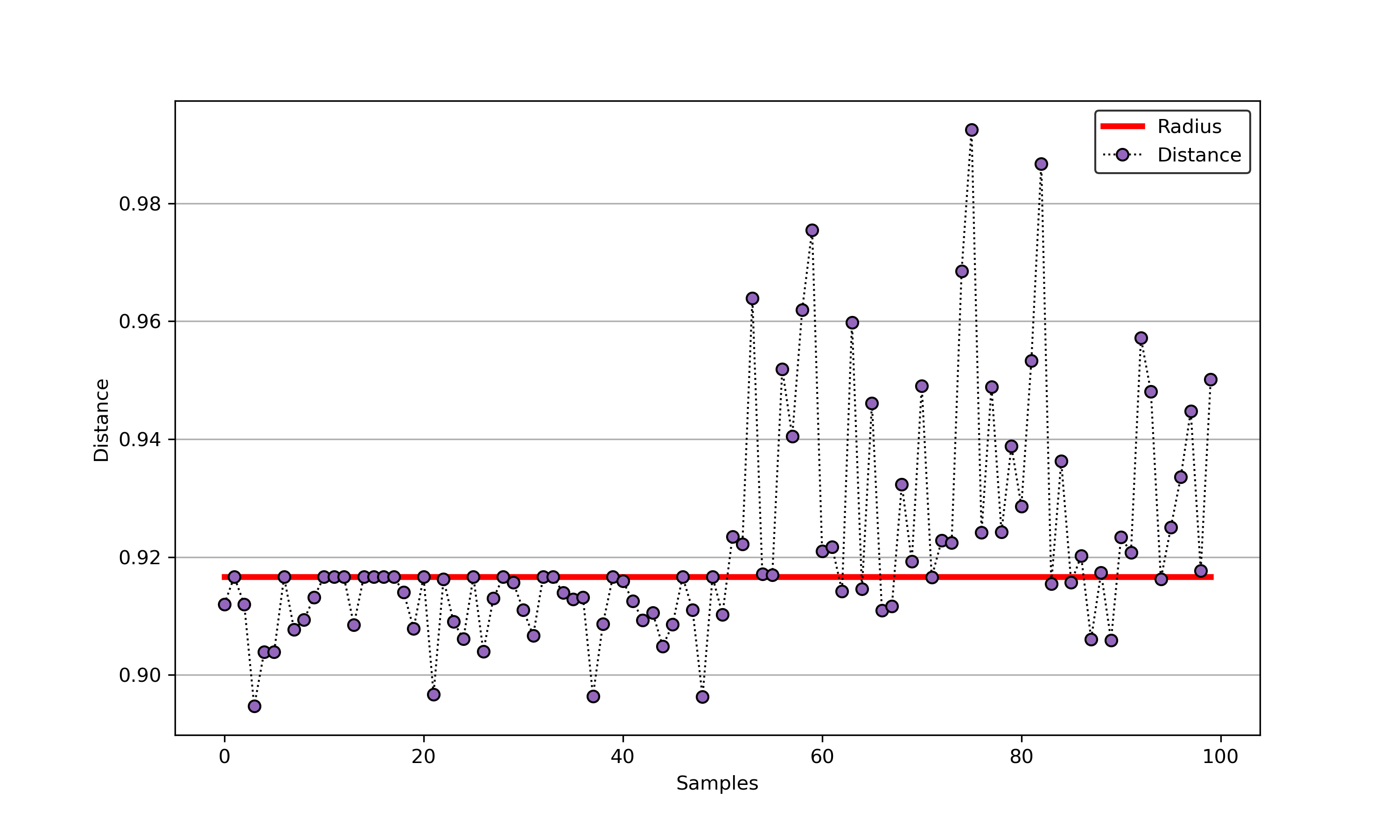
An example for SVDD model fitting with negataive samples.
import syssys.path.append("..")from sklearn.datasets import load_winefrom src.BaseSVDD import BaseSVDD, BananaDataset# Banana-shaped dataset generation and partitioningX, y = BananaDataset.generate(number=100, display='on')X_train, X_test, y_train, y_test = BananaDataset.split(X, y, ratio=0.3)#svdd = BaseSVDD(C=0.9, gamma=0.3, kernel='rbf', display='on')#svdd.fit(X_train, y_train)#svdd.plot_boundary(X_train, y_train)#y_test_predict = svdd.predict(X_test, y_test)#radius = svdd.radiusdistance = svdd.get_distance(X_test)svdd.plot_distance(radius, distance)
An example for SVDD model fitting using different kernels.
import syssys.path.append("..")from src.BaseSVDD import BaseSVDD, BananaDataset# Banana-shaped dataset generation and partitioningX, y = BananaDataset.generate(number=100, display='on')X_train, X_test, y_train, y_test = BananaDataset.split(X, y, ratio=0.3)# kernel listkernelList = {"1": BaseSVDD(C=0.9, kernel='rbf', gamma=0.3, display='on'),"2": BaseSVDD(C=0.9, kernel='poly',degree=2, display='on'),"3": BaseSVDD(C=0.9, kernel='linear', display='on')}#for i in range(len(kernelList)):svdd = kernelList.get(str(i+1))svdd.fit(X_train, y_train)svdd.plot_boundary(X_train, y_train)

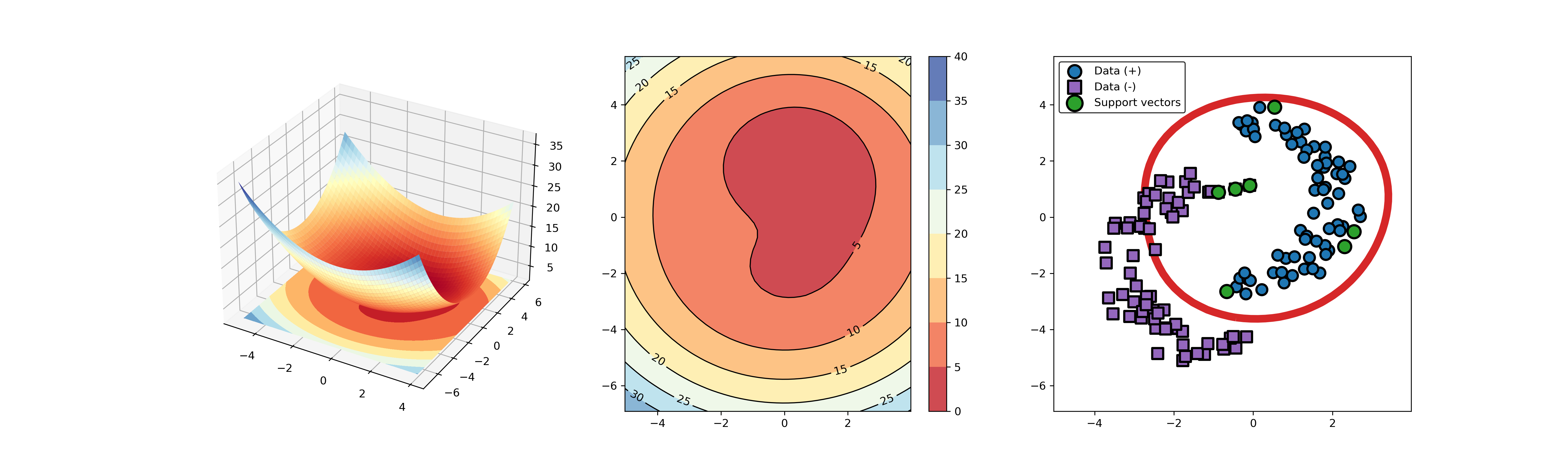
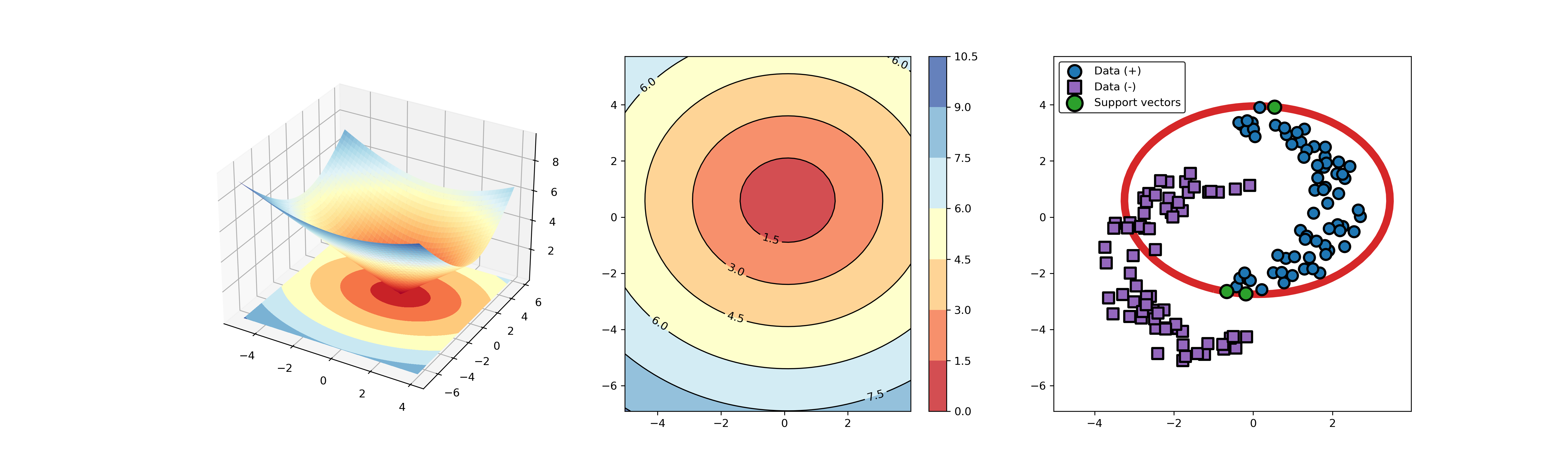
An example for SVDD model fitting using nonlinear principal component.
The KPCA algorithm is used to reduce the dimension of the original data.
import syssys.path.append("..")import numpy as npfrom src.BaseSVDD import BaseSVDDfrom sklearn.decomposition import KernelPCA# create 100 points with 5 dimensionsX = np.r_[np.random.randn(50, 5) + 1, np.random.randn(50, 5)]y = np.append(np.ones((50, 1), dtype=np.int64),-np.ones((50, 1), dtype=np.int64),axis=0)# number of the dimensionalitykpca = KernelPCA(n_components=2, kernel="rbf", gamma=0.1, fit_inverse_transform=True)X_kpca = kpca.fit_transform(X)# fit the SVDD modelsvdd = BaseSVDD(C=0.9, gamma=10, kernel='rbf', display='on')# fit and predictsvdd.fit(X_kpca, y)y_test_predict = svdd.predict(X_kpca, y)# plot the distance curveradius = svdd.radiusdistance = svdd.get_distance(X_kpca)svdd.plot_distance(radius, distance)# plot the boundarysvdd.plot_boundary(X_kpca, y)
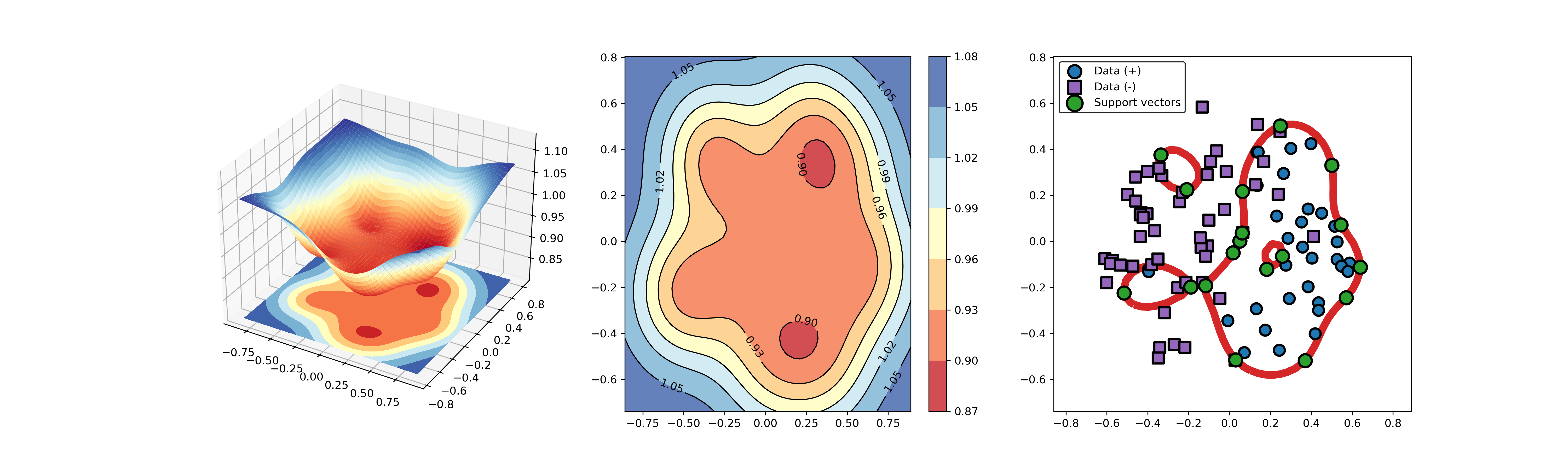
An example for parameter optimization using PSO.
“scikit-opt” is required in this example.
https://github.com/guofei9987/scikit-opt
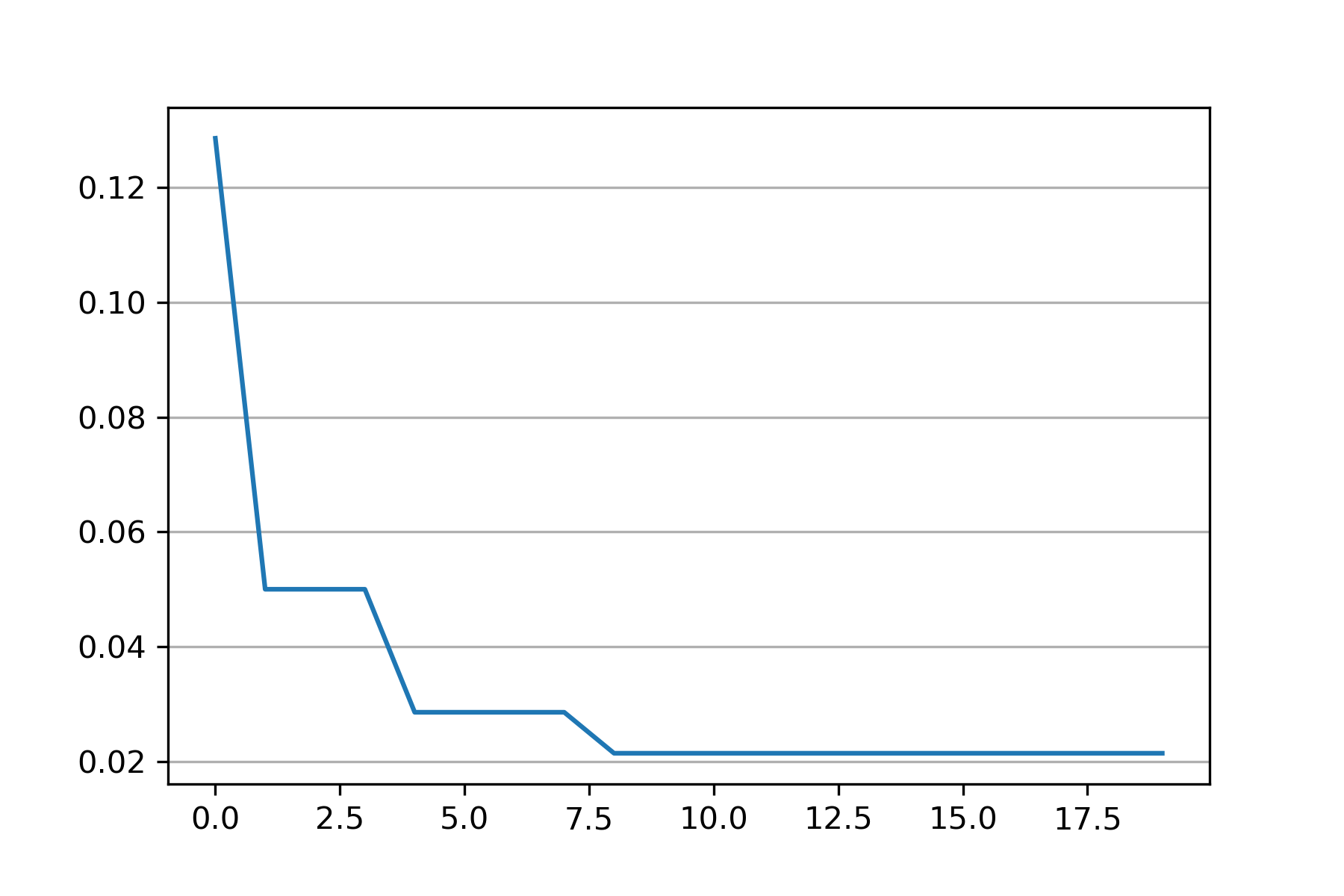
import syssys.path.append("..")from src.BaseSVDD import BaseSVDD, BananaDatasetfrom sko.PSO import PSOimport matplotlib.pyplot as plt# Banana-shaped dataset generation and partitioningX, y = BananaDataset.generate(number=100, display='off')X_train, X_test, y_train, y_test = BananaDataset.split(X, y, ratio=0.3)# objective functiondef objective_func(x):x1, x2 = xsvdd = BaseSVDD(C=x1, gamma=x2, kernel='rbf', display='off')y = 1-svdd.fit(X_train, y_train).accuracyreturn y# Do PSOpso = PSO(func=objective_func, n_dim=2, pop=10, max_iter=20,lb=[0.01, 0.01], ub=[1, 3], w=0.8, c1=0.5, c2=0.5)pso.run()print('best_x is', pso.gbest_x)print('best_y is', pso.gbest_y)# plot the resultfig = plt.figure(figsize=(6, 4))ax = fig.add_subplot(1, 1, 1)ax.plot(pso.gbest_y_hist)ax.yaxis.grid()plt.show()
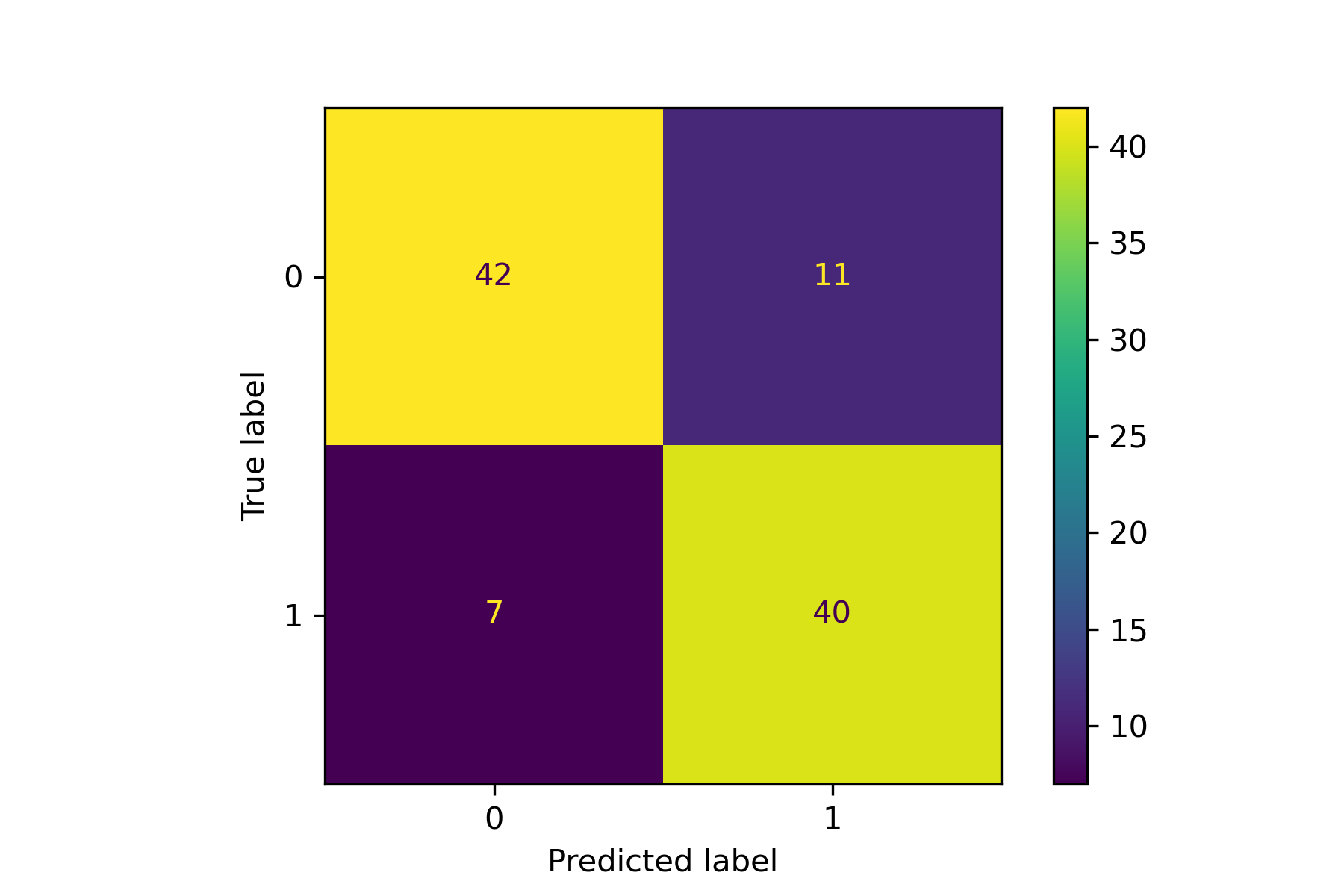
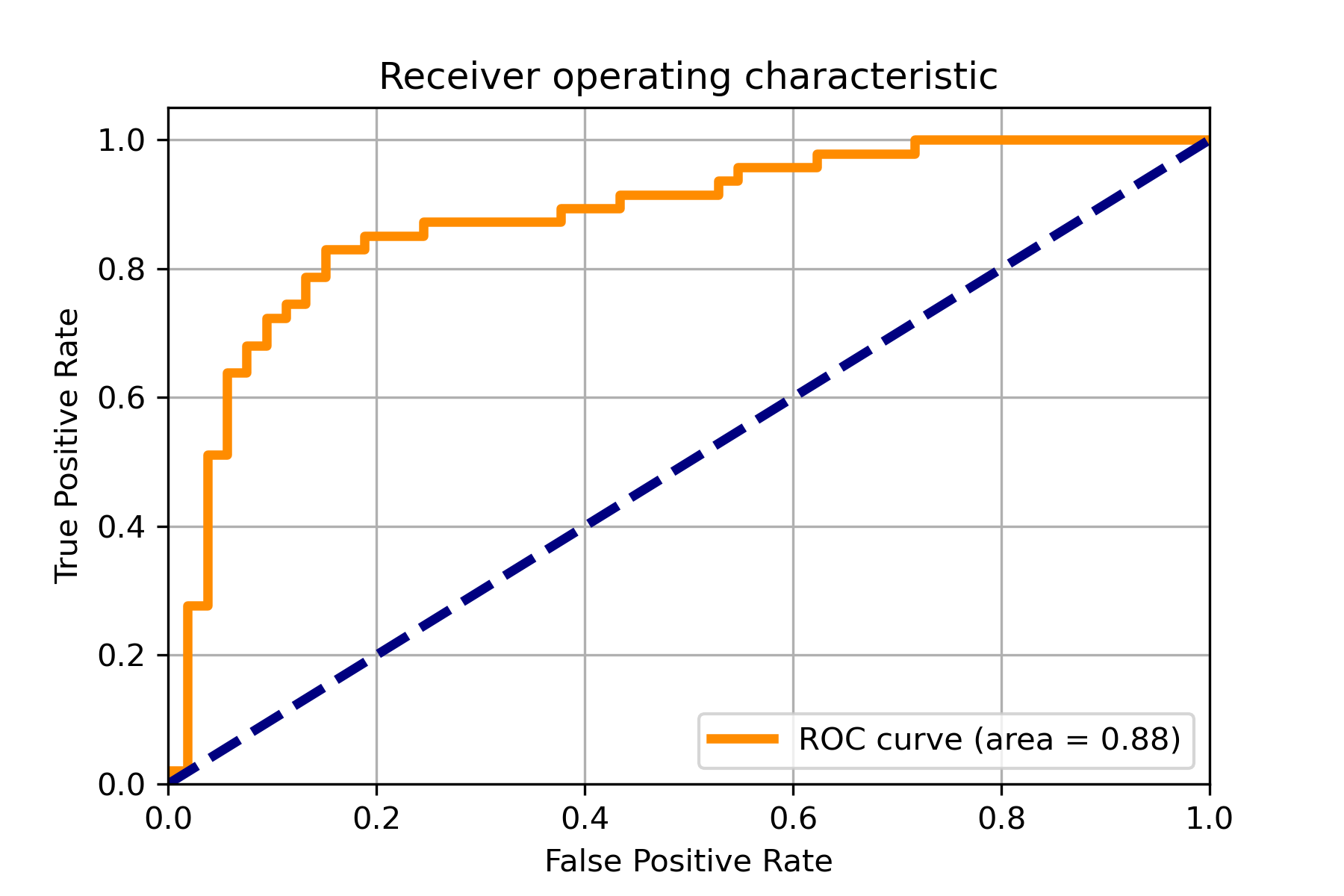
An example for drawing the confusion matrix and ROC curve.
An example for cross validation.
import syssys.path.append("..")from src.BaseSVDD import BaseSVDD, BananaDatasetfrom sklearn.model_selection import cross_val_score# Banana-shaped dataset generation and partitioningX, y = BananaDataset.generate(number=100, display='on')X_train, X_test, y_train, y_test = BananaDataset.split(X, y, ratio=0.3)#svdd = BaseSVDD(C=0.9, gamma=0.3, kernel='rbf', display='on')# cross validation (k-fold)k = 5scores = cross_val_score(svdd, X_train, y_train, cv=k, scoring='accuracy')#print("Cross validation scores:")for scores_ in scores:print(scores_)print("Mean cross validation score: {:4f}".format(scores.mean()))
Results
Cross validation scores:0.57142857142857140.750.96428571428571431.01.0Mean cross validation score: 0.857143
An example for parameter selection using grid search.
import syssys.path.append("..")from sklearn.datasets import load_winefrom src.BaseSVDD import BaseSVDD, BananaDatasetfrom sklearn.model_selection import KFold, LeaveOneOut, ShuffleSplitfrom sklearn.model_selection import learning_curve, GridSearchCV# Banana-shaped dataset generation and partitioningX, y = BananaDataset.generate(number=100, display='off')X_train, X_test, y_train, y_test = BananaDataset.split(X, y, ratio=0.3)param_grid = [{"kernel": ["rbf"], "gamma": [0.1, 0.2, 0.5], "C": [0.1, 0.5, 1]},{"kernel": ["linear"], "C": [0.1, 0.5, 1]},{"kernel": ["poly"], "C": [0.1, 0.5, 1], "degree": [2, 3, 4, 5]},]svdd = GridSearchCV(BaseSVDD(display='off'), param_grid, cv=5, scoring="accuracy")svdd.fit(X_train, y_train)print("best parameters:")print(svdd.best_params_)print("\n")#best_model = svdd.best_estimator_means = svdd.cv_results_["mean_test_score"]stds = svdd.cv_results_["std_test_score"]for mean, std, params in zip(means, stds, svdd.cv_results_["params"]):print("%0.3f (+/-%0.03f) for %r" % (mean, std * 2, params))print()
Results
best parameters:{'C': 0.5, 'gamma': 0.1, 'kernel': 'rbf'}0.921 (+/-0.159) for {'C': 0.1, 'gamma': 0.1, 'kernel': 'rbf'}0.893 (+/-0.192) for {'C': 0.1, 'gamma': 0.2, 'kernel': 'rbf'}0.857 (+/-0.296) for {'C': 0.1, 'gamma': 0.5, 'kernel': 'rbf'}0.950 (+/-0.086) for {'C': 0.5, 'gamma': 0.1, 'kernel': 'rbf'}0.921 (+/-0.131) for {'C': 0.5, 'gamma': 0.2, 'kernel': 'rbf'}0.864 (+/-0.273) for {'C': 0.5, 'gamma': 0.5, 'kernel': 'rbf'}0.950 (+/-0.086) for {'C': 1, 'gamma': 0.1, 'kernel': 'rbf'}0.921 (+/-0.131) for {'C': 1, 'gamma': 0.2, 'kernel': 'rbf'}0.864 (+/-0.273) for {'C': 1, 'gamma': 0.5, 'kernel': 'rbf'}0.807 (+/-0.246) for {'C': 0.1, 'kernel': 'linear'}0.821 (+/-0.278) for {'C': 0.5, 'kernel': 'linear'}0.793 (+/-0.273) for {'C': 1, 'kernel': 'linear'}0.879 (+/-0.184) for {'C': 0.1, 'degree': 2, 'kernel': 'poly'}0.836 (+/-0.305) for {'C': 0.1, 'degree': 3, 'kernel': 'poly'}0.771 (+/-0.416) for {'C': 0.1, 'degree': 4, 'kernel': 'poly'}0.757 (+/-0.448) for {'C': 0.1, 'degree': 5, 'kernel': 'poly'}0.871 (+/-0.224) for {'C': 0.5, 'degree': 2, 'kernel': 'poly'}0.814 (+/-0.311) for {'C': 0.5, 'degree': 3, 'kernel': 'poly'}0.800 (+/-0.390) for {'C': 0.5, 'degree': 4, 'kernel': 'poly'}0.764 (+/-0.432) for {'C': 0.5, 'degree': 5, 'kernel': 'poly'}0.871 (+/-0.224) for {'C': 1, 'degree': 2, 'kernel': 'poly'}0.850 (+/-0.294) for {'C': 1, 'degree': 3, 'kernel': 'poly'}0.800 (+/-0.390) for {'C': 1, 'degree': 4, 'kernel': 'poly'}0.771 (+/-0.416) for {'C': 1, 'degree': 5, 'kernel': 'poly'}